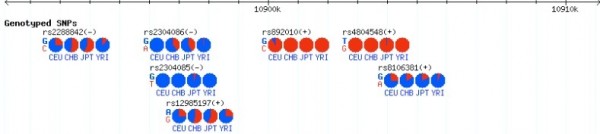
基因组的某位置,会发生突变,或者说对于多个物种的个体进行测序的话,会发现许多位置不是稳定的只是某个碱基,而应该用频率表示,ACTG出现的频率,如果某个位置有出现两个以上碱基的可能性,我们就将这个位置称为SNV,现在GBrowse中展示这些SNV,不仅是其位置,还要展示各个碱基出现的频率。
思路1,适用 glyph=image
根据Bio::Graphics中对于image的解释:可以传入一个url或者本地的路径,我想能不能创建一个动态画图的URL,其可以根据传入的参数,生成一个饼状的比例图,然后再配置文件中,动态生成image的url地址,如下:
image = http://localhost/cgi-bin/snp.pl?note=$description link = http://localhost/cgi-bin/snp.pl?note=$descriptio
结果:没有显示频率图,可能是传入参数不正确,也就是link可用的变量,对于image是无效的,查阅Bio::Graphics::Glyph::image后,修改配置
image = sub{my $feature = shift;
my $img_path="http://localhost/cgi-bin/snp.pl?note=" . $feature->desc;
return $img_path;}
link = http://localhost/cgi-bin/snp.pl?note=$description结果:没有图像显示,怀疑image不支持callbake,将该sub用于link,有效,只是description没有显示全,修改配置:
image = sub {
my $f = shift;
return 'http://localhost/cgi-bin/snp.pl?note=' . $f->attributes('Note');
}结果依然,没有图片显示。
思路2:Bio::Graphics::Glyph::allele_tower
http://search.cpan.org/~lds/Bio-Graphics-2.21/lib/Bio/Graphics/Glyph/allele_tower.pm
似乎只可以显示都有什么基因型
进一步思路
- 是否有其他类型支持
- glyph是否支持callbake,进行画图
- 文字显示description = 1
- 冒泡提示显示
- 使用插件
后来进一步的搜索,还是可以找到这样的插件:
Hapmap uses pie charts to show allele frequency information for the 4
populations. These are rendered by a plugin allele_pie_multi.pm,
downloadable from:
http://www.hapmap.org/downloads/gbrowse/2005-03_phaseI/glyph/allele_pie_multi.pm
Copy this into the gbrowse.conf/plugins directory.
I use the following (modified) configuration:
[WILL_TEST]
feature = method:source
glyph = allele_pie_multi
ref_allele = sub {return shift->attributes('Refallele')}
freq = sub {
my $snp = shift;
my @GTcounts = sort $snp->attributes('GTcounts');
my %freqs;
my $refallele = $snp->attributes('Refallele');
foreach(@GTcounts) {
my ($cohort, $data) = split(/:/);
my %pairs = split(/[= ]/, $data);
$freqs{$cohort} = $pairs{$refallele};
}
# If there's just one cohort, don't display its name
if (keys %freqs == 1) {
my $cohort = (keys %freqs)[0];
%freqs = ( '' => $freqs{$cohort} );
}
return join ';', map { exists $freqs{$_} ?
"$_:$freqs{$_}" : "$_:NO" } sort keys %freqs;
}
alleles = sub {return shift->attributes('Alleles')}
ref_strand = sub {shift->strand}
font2color = #0000FF
bgcolor = red
#stacked = 1
label density = 50
bump density = 200
key = Will test
link = sub {
my $feature=shift;
my $dbsnp_id=$feature->name;
$dbsnp_id=~s/rsrs/rs/;
return
"http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=$dbsnp_id" if $dbsnp_id;
return 'wibble';
}
description = 1
label = sub{ my $self = shift;
my $s = $self->strand;
my $n = $self->name;
return $n if $s == 0;
my $m = "+" if $s > 0;
$m = "-" if $s < 0;
return $n . "(" . $m . ")";
}This will handle gff data loaded with the following format:
chr1 source method 792429 792429 0.968 – . Probe rs3094315 ; Alleles A/G ; Refallele A ; GTcounts “coh1:A=0.84 G=0.160″ ; GTcounts “coh2:A=0.837 G=0.163″
The plugin was written to handle allele frequencies, for two alleles, and can only render the pies binarily. You would have to adapt the plugin to get frequencies displayed for all 4 alleles.
本文来自:http://boyun.sh.cn/bio/?p=1823