The single file I’ll discuss today has in fact almost the entire assembly in it, besides the actual sequences (although even some of these are also included, see below). As explained in my first post, newbler (as many other assembly programs) builds a contig graph. Contigs are the nodes, and reads spanning between them (starting in one contig and continuing or ending in another) indicate the edges. All the information on this graph, except the actual read alignments and consensus contigs, is in the 454ContigGraph.txt file.
The file is divided into several sections, for each one the lines start with a capital letter, except for the first section.
Section 1) Contig statistics
1 contig00001 27007 29.7
2 contig00002 34455 29.1
3 contig00003 35840 28.0
4 contig00004 32644 30.0
5 contig00005 20873 27.8
This first part consists, for each contig, of
- the number (identifier) of the contig
- the contig name (‘contig’ followed by the contig number with zeros so that it has at least 5 digits)
- the length of the contig
- its read depth.
‘Read depth’ is defined as the total number of included bases from all the reads aligned to generate the consensus contig sequence, divided over the contig length. Note that reads at the ends of contigs can contribute as little as 1 base (yes, I’ve seen it) to the contig. In that case, only that single base is counted towards the total. Most reads, however, count over the entire length.
This section is the only one where the contigs are named with the actual word ‘contig’. In the remainder of the file, the contig number are used.
Section 2) Edges (lines starting with ‘C’)
C 26 3' 221 3' 24
C 27 3' 1754 5' 23
C 27 3' 62 5' 22
C 212 5' 1924 3' 27
C 21 5' 1034 5' 21
C 28 5' 1034 5' 25
C 29 3' 1895 3' 45
C 127 5' 31 3' 32
Edges are reads that align from the end (5’ or 3’) of one contig into the end (5’ or 3’) of another. The columns represent:
- the letter ‘C’
- the contig number on the left end of the edge
- 5’ or 3’ to indicate which end of the contig the left edge refers to
- the contig number at the right end of the edge
- 5’ or 3’ to indicate which end of the contig the right edge refers to
- the depth of the edge.
‘Edge depth’ represents the number of reads that align to form this edge.
From the example it already becomes clear that the graph can be complicated: contig 27 has two 3’ edges. From the first section of the file, it appears that its depth is also about twice the average, indicating that contig 27 most likely is a collapsed repeat (see the post on how newbler works). The same holds for contig 1034, which has about triple the average depth, and three 5’ edges (the third one is not shown in the small excerpt).
Section 3) Scaffolds (lines starting with ‘S’)
S 21 249301 1048:+;gap:1209;1049:+;gap:222;1050:+;gap:1329;1051:+;gap:721;1052:+;gap:729;1053:+;gap:542;1054:+;gap:807;1055:+;gap:305;1056:+;gap:644;1057:+
The information in this section is partly overlapping with that in the 454Scaffolds.txt file (described in the previous post). The first three columns are:
- the letter ‘S’
- the scaffold number (this would make this scaffold00021)
- the scaffold length
The last column describes how the scaffold is built up out of contigs and gaps:
1048:+;gap:1209;1049:+;gap:222;1050:+;gap:1329;1051:+
means that scaffold00021
- starts with contig 1048 (contig01048), in the + (forward) orientation (i.e. 5’ to 3’)
- followed by a gap of 1209 bp
- followed by contig 1049
- a 222 bp gap
- contig 1050
- etc…
Section 4) Thru-flow information (lines starting with ‘I’)
I 34 TCTTATAAAGAAACGGTTTATTATATAAGTAGTATCTGGGAAAAGGCAGATTTTTTTTCCCAAAAGATTAAAGGGCATTGGG 15:1805-3'..207-3';14:1805-3'..1973-5'
I 35 AACTTTTCCTCCGTAAATACCGTTAATGTTTCTGGAAATTCAGTTACATTAGACACCAGTATTGGAAATGGAGCAATTGACTTTATTGGTTCAACCCTTGCTGGA 10:36-3'..36-5'
I 91 ACCACTTATTTCGA 85:93-5'..92-5'
For (very) short contigs that consist of short repeats, reads that have that repeat in them are often longer than the repeat/contig length. In this case, the reads would ‘flow through’ the short contig, starting in a contig outside it, and ending in yet another contig. This section has an entry for all contigs shorter than 256 bp, and if there are flow-through reads, some statistics on these are included. The columns are:
- The letter ‘I’
- The contig number
- The contig sequence (note, this only included if the contig has less than 256 columns in the alignment)
- The ‘Thru-flow’ information
In the example for contig 34 (or contig00034):
15:1805-3'..207-3';14:1805-3'..1973-5'
This means that
- there are 15 reads that come from the 3’ end of contig 1805, flow through contig 34, and continue at the 3’ end of contig 207, AND
- there are 14 reads that start come from the 3’ end of contig 1805, flow through contig 34, and continue at the 5’ end of contig 1973.
Section 5) Single-end Read flow information (lines starting with ‘F’)
F 3 1033/40/0.0 1851/57/36.1;1808/41/68.3
F 4 124/67/0.0 117/46/0.0;5/3/101.3
F 5 - 1008/31/0.0
This section contains information on where reads end (or start) that flow out of the contig in question (i.e. have their start or end in the contig in question, but do not align entirely in it). The columns are:
- The letter ‘F’
- The contig number
- The flow information for reads flowing from the 5’ end of the contig
- The flow information for reads flowing from the 3’ end of the contig
In the example for contig 4 (or contig00004):
124/67/0.0 117/46/0.0;5/3/101.3
- For the 5’ flows: 67 reads flow from the 5’ end of contig 4 and terminate in contig 124; the average distance from the 5′ end of contig 4 to the end of contig 124 into which the reads flow is 0.0 bp. Zero bp? Yes, this means that the two contigs are right next to each other without a gap inbetween. Zero base gaps between contigs are logical if you understand the contig graph principle: collapsed repeat contigs branch off on either end into single-copy contigs. People who first start mining newbler assemblies are sometimes frustrated to find contigs that seem to belong next to each other without a gap…
- For the 3’ flows: 46 reads flow from the 3’ end of contig 4 and terminate in contig 117; again, these contigs are next to each other.
- An additional 3 reads flow from the 3’ end of contig 4 and terminate in contig 5; the average distance from the 3′ end of contig 4 to the end of contig 5 into which the reads flow is 101.3 bp. Contig 117 is 101 bp, indicating that the reads most likely flow through this contig!
Note that the ‘minus’ for the 5’ flows of contig 5 in the excerpt above indicates there are no such flows for this contig.
Section 6) Paired-end Read flow information (lines starting with ‘P’)
This section describes essentially the same information as the previous one, except that it deals with the paired end reads.
As you can see, this file contains a lot of information. What use is it? One immediate use is the read depth described in the first section. When you plot the distribution of read depths, you get a feel for the overall coverage (‘oversampling’) of the assembly. Also, contigs with unusually low depth could indicate contamination, those with unusually high depth collapsed repeats. In fact, read depth turns out to be correlated to the number of copies present in the genome, a fact that my colleagues and I exploited in a paper available here.
Also, the information about which contigs are gapless neighbors could come in handy.
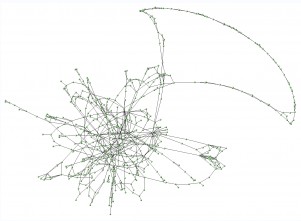
In addition, I think it could be very useful if there would exist a browser for the contig graph. It would allow for looking visually at neighboring contigs, indicate which contigs are repeated and where these could be placed, explain gaps in scaffolds etc. I once used a very simple approach, treating contigs as ‘dots’, disregarding the 5’ and 3’ ends, contig length and depth, only indicating contig edges, and made a graph in VisANT. This program will accept a simple table describing ‘from to ‘ and direction (+1 or -1, which I set to +1 for all). A nice feature of VisANT is that it allows for determining all possibleshortests paths between selected nodes. I used it to check if certain contigs could be (close) neighbors. This is a VisANT example for a bacterial genome assembly :