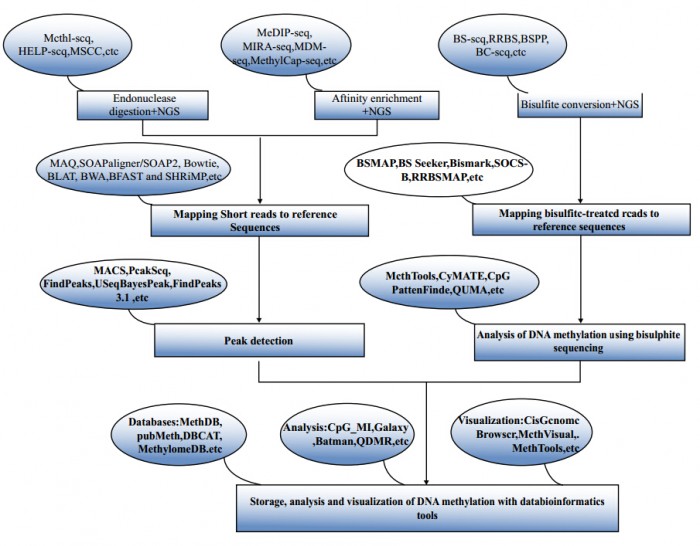
DNA甲基化分析流程示意图
Workflow of high-throughput DNA methylation analysis. The workflow shows the basic process of high-throughput DNA methylation analysis by existing bioinformatics tools. The bioinformatics tools are shown in the ellipses and the corresponding functions in the neighboring boxs.
表1:基于二代测序检测DNA甲基化的方法(NGS-based technologies for detecting DNA methylation)
| Pretreatment method | Genome coverage | NGS-based analysis | Application | Ref |
| Endonuclease digestion | Moderate | Methl-seq | Assay a range of genomic elements; allowing a broader survey of regions than classic methylation studies limited to CpG islands and promoters | [16] |
| HELP-seq | Measurement of repetitive sequences, copy-number variability, allele-specific and smaller fragments (<50bp) ;Sensitivity of detection of hypomethylated loci | [12,75] | ||
| MSCC | Identification of the unmethylated region of a genome by pinpointing unmethylated CpGs at single base-pair resolution | [13,14] | ||
| Affinity enrichment | Moderate | MeDIP-seq | Generation of unbiased, cost-effective, and full-genome methylation levels without the limitations of restriction sites or CpG islands; | [1,76] |
| MIRA-seq | Analyze recovered or double-stranded methylated DNA on a genome-wide scale; Applicable to various clinical and diagnostic situations. | [19] | ||
| MDB-seq | Applied to any biological settings to identify differentially methylated regions at the genomic scale | [18] | ||
| MethylCap-seq | Detection of differentially methylated regions with high genome coverage; Detect DMRs in clinical samples | [20,77] | ||
| Bisulfite conversion | High | BS-seq | Sensitively measure cytosine methylation on a genome-wide scale within specific sequence contexts | [48,78] |
| RRBS | Analyze a limited number of gene promoters and regulatory sequence elements in a large number of samples; Analyzing and comparing genomic methylation patterns | [11,28] | ||
| BSPP | Focus sequencing on the most informative genomic regions ;exon capturing and SNP genotyping; Detecting methylation in large genomes | [29] | ||
| BC-seq | Detect site-specific switches in methylation; Determine DNA methylation frequencies in CGIs sampled from a variety of genomic settings including promoters, exons, introns, and intergenic loci | [30] |
表2:短Reads比对算法(Alignment tools for short reads.)
| Resources | URL | Ref |
| MAQ | http://maq.sourceforge.net/ | [33] |
| SOAPaligner/soap2 | http://soap.genomics.org.cn/index.html | [36] |
| Bowtie | http://bowtie-bio.sourceforge.net/index.shtml | [32] |
| BLAT | http://genome.ucsc.edu/cgi-bin/hgBlat | [79] |
| BWA | http://maq.sourceforge.net/ | [80] |
| BFAST | http://bfast.sourceforge.net | [81] |
| SHRiMP | http://compbio.cs.toronto.edu/shrimp. | [82] |
备注:
- SOAP, Short Oligonucleotide Analysis Package;
- MAQ, Mapping and Assembly with Quality;
- BLAT, BLAST-like alignment tool;
- BWA, Burrows-Wheeler Alignment;
- BFAST, BLAT-like Fast Accurate Search Tool;
- SHRiMP, The Short-Read Mapping Package.
表3:检测峰值算法( Peak detection algorithms)
| Algorithm | Description | Availability | Ref |
| MACS | Model-based analysis of ChIP-Seq | http://liulab.dfci.harvard.edu/MACS/ | [41] |
| ChIPseeqer | in-depth analysis of ChIP-seq datasets | http://physiology.med.cornell.edu/faculty/elemento/lab/CS_files/ChIPseeqer-2.0.tar.gz | [83] |
| HPeak | A HMM-based algorithm for defining read enriched regions | www.sph.umich.edu/csg/qin/HPeak | [84] |
| CASSys | ChIP-seq data Analysis Software System | http://localness.zbh.uni-hamburg.de/~ProjektChipSeq/cgi-bin/login.rb | [85] |
| PeakSeq | A general scoring approach for ChIP-seq data analysis. | http://info.gersteinlab.org/PeakSeq | [86] |
| Sole-Search | Integrated peak-calling and analysis software | http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi | [87] |
| SISSRS | Precise identification of binding sites from short reads generated from ChIP-Seq experiment | http://sissrs.rajajothi.com/ | [88] |
| BayesPeak | Bayesian analysis of ChIP-seq data | http://bioconductor.org/packages/release/bioc/html/BayesPeak.html | [42] |
| PeakRanger | A cloud-enabled peak caller for ChIP-seq data | http://www.modencode.org/software/ranger/ | [89] |
| FindPeaks 3.1 | A tool for identifying areas of enrichment | http://www.bcgsc.ca/platform/bioinfo/software/findpeaks | [40] |
| Sole-Search | An integrated analysis program for peak detection and functional annotation using ChIP-seq data | http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi | [87] |
| PeakAnalyzer | Genome-wide annotation of chromatin binding and modification loci | http://www.bioinformatics.org/peakanalyzer/wiki/ | [90] |
| chromaSig | A Probabilistic Approach to Finding Common Chromatin Signatures | http://bioinformatics-renlab.ucsd.edu/rentrac/wiki/ChromaSig | [43] |
| Fish the ChIPs | A pipeline for automated genomic annotation of ChIP-Seq data | http://bio.ifom-ieo-campus.it/ftc/ | [91] |
表4:分析亚硫酸盐测序测序数据的计算工具(Analysis of bisulfite sequencing data with computational tools)
| Resources | Purpose | URL | Ref |
| RRBSMAP | A fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing | http://rrbsmap.computational-epigenetics.org/ | [92] |
| BSMAP | Ahole genome bisulfite sequence mapping | http://code.google.com/p/bsmap/ | [48] |
| BS Seeker | Precise mapping for bisulfite sequencing | http://pellegrini.mcdb.ucla.edu/BS_Seeker/BS_Seeker.html | [46] |
| Bismark | Map and determine the Methylation state of BS-Seq read | http://www.bioinformatics.bbsrc.ac.uk/projects/bismark/ | [47] |
| SOCS-B | An alignment algorithm for bisulfite sequencing using the Applied Biosystems SOLiD System | http://solidsoftwaretools.com/gf/project/socs/ | [93] |
| BRAT | Bisulfite-treated reads analysis tool | http://compbio.cs.ucr.edu/brat/ | [94] |
| BISMA | Analysis of bisulfite Sequencing data from both unique and repetitive sequences | http://biochem.jacobs-university.de/BDPC/BISMA/ | [95] |
| BiQAnalyzerHT | Locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing | http://biq-analyzer-ht.bioinf.mpi-inf.mpg.de/ | [49] |
| CpGviewer | Sequence analysis and editing for bisulphite genomic sequencing projects | http://xserve1.leeds.ac.uk/~iancarr/cpgviewer | [50] |
| CpG PatternFinder | Windows-based program for bisulphite DNA | - | [52] |
| CyMATE | Bisulphite-based analysis of plant genomic DNA | http://www.gmi.oeaw.ac.at/CyMATE | [51] |
| GenomeStudio Software | Analyzing data generated from Illumina assays | - | - |
| MethMarker | Design, optimize and validate DNA methylation biomarkers for a given DMR | http://methmarker.mpi-inf.mpg.de/ | [96] |
| BDPC | Bisulfite sequencing Data methylation analysis. | http://biochem.jacobs-university.de/BDPC | [97] |
| MethylCoder | Software pipeline for bisulte-treated sequences | https://github.com/brentp/methylcode | [98] |
| QUMA | Quantification tool for methylation analysis | http://quma.cdb.riken.jp/ | [53] |
备注:
- CyMATE ,Cytosine Methylation Analysis Tool for Everyone;
- BSMAP, Bisulphite Sequence Mapping Program;
- BISMA, Bisulfite Sequencing DNA Methylation Analysis;
- BRAT,bisulte-treated reads analysis tool;
- DMR, differentially methylated DNA region;
- QUMA, Quantification Tool For Methylation Analysis.
表5:其他生物信息学软件(Bioinformatics tools)
| Tools | Purpose | UTR | Ref |
| MethDB | Database for DNA methylation data | http://www.methdb.de | [54] |
| MethyCancer Database | Database of cancer DNA methylation data | http://methycancer.psych.ac.cn/ | [57] |
| PubMeth | Database of DNA methylation literature | http://www.pubmeth.org/ | [99] |
| NGSmethDB | Database for DNA methylation data at single-base resolution | http://bioinfo2.ugr.es/NGSmethDB/gbrowse/ | [55] |
| DBCAT | Database of CpG islands and analytical tools for identifying comprehensive methylation profiles in cancer cells | http://dbcat.cgm.ntu.edu.tw/ | [100] |
| MethylomeDB | Database of DNA methylation profiles of the brain | http://epigenomics.columbia.edu/methylomedb/index.html | [56] |
| DiseaseMeth | Human disease methylation database | http://bioinfo.hrbmu.edu.cn/diseasemeth | [58] |
| CpG IE | Identification of CpG islands | http://bioinfo.hku.hk/cpgieintro.html | [63] |
| CpG IS | Identification of CpG islands | http://cpgislands.usc.edu/ | [64] |
| CG clusters | Identification of CpG islands | http://greallylab.aecom.yu.edu/cgClusters/ | [68] |
| CpGcluster | Identification of CpG islands | http://bioinfo2.ugr.es/CpGcluster | [67] |
| CpGIF | Identification of CpG islands | http://www.usd.edu/~sye/cpgisland/CpGIF.htm | [101] |
| CpG_MI | Identification of CpG islands | http://bioinfo.hrbmu.edu.cn/cpgmi | [66] |
| CpGProD | Identification of CpG islands | http://pbil.univ-lyon1.fr/software/cpgprod.html | [65] |
| EpiGRAPH | Genome scale statistical analysis | http://epigraph.mpi-inf.mpg.de/WebGRAPH | [71] |
| Galaxy | General purpose analysis | http://main.g2.bx.psu.edu/ | [102] |
| QDMR | Identification of differentially methylated regions | http://bioinfo.hrbmu.edu.cn/qdmr. | [103] |
| Batman | MeDIP DNA methylation analysis tool | http://td-blade.gurdon.cam.ac.uk/software/batman | [23] |
| CisGenome Browser | A flexible tool for genomic data visualization | http://biogibbs.stanford.edu/~jiangh/browser/ | [70] |
| MethVisual | Visualization and exploratory statistical analysis of DNA methylation profiles from bisulfite sequencing | http://methvisual.molgen.mpg.de/ | [104] |
| MethTools | A toolbox to visualize and analyze DNA methylation data | http://genome.imb-jena.de/methtools/ | [105] |
以上内容摘选自:Su J, Huang D, Yan H, Liu H, Zhang Y (2012) Advances in Bioinformatics Tools for High-Throughput Sequencing Data of DNA Methylation. Hereditary Genet 1:107. doi: 10.4172/2161-1041.1000107。
各个软件后的参考文献请阅读论文原文查找。