基于RNA-Seq的转录组数据分析已经在研究中运用了近10来年了,现在一些杂志在发表论文的时候reviewers已经倾向于用RNA-Seq来替代RT-qPCR。对于生物信息专业“干实验”的同学来说,RNA-Seq数据分析可以说是信手拈来,但是对于专注于“湿实验”的同学来说,RNA-Seq听起来似乎是那么近但是操作起来似乎又好远。我身边也经常有专注于“湿实验”的朋友经常来问我各种关于RNA-Seq的信息。为了方便更多同学了解活着深入系统地了解RNA-Seq。这里分享一篇review,这篇主要侧重于理论方面,给大家普及介绍一下RNA-Seq相关知识,算是扫盲吧。
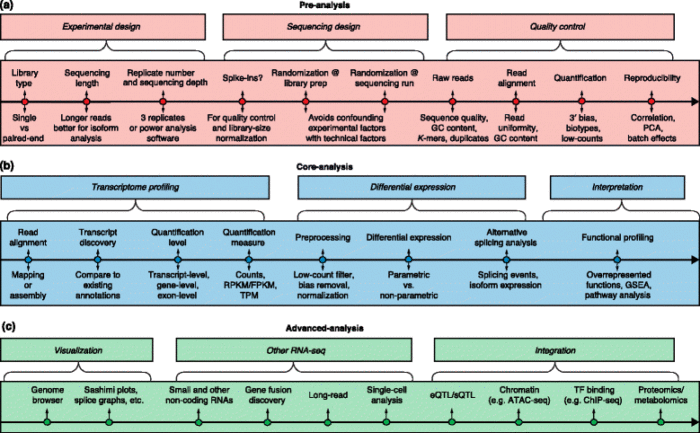
下面这张图主要介绍RNA-Seq都有哪些分析可以开展。
更多细节的介绍还是阅读原文吧。
A survey of best practices for RNA-seq data analysis
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-0881-8
RNA-sequencing (RNA-seq) has a wide variety of applications, but no single analysis pipeline can be used in all cases. We review all of the major steps in RNA-seq data analysis, including experimental design, quality control, read alignment, quantification of gene and transcript levels, visualization, differential gene expression, alternative splicing, functional analysis, gene fusion detection and eQTL mapping. We highlight the challenges associated with each step. We discuss the analysis of small RNAs and the integration of RNA-seq with other functional genomics techniques. Finally, we discuss the outlook for novel technologies that are changing the state of the art in transcriptomics.