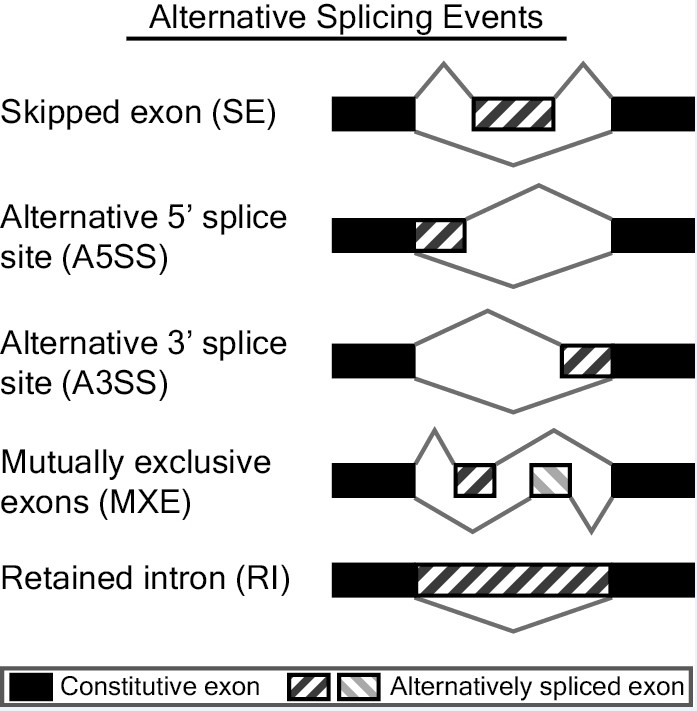
给大家推荐一款最新在Nucleic Acids Research上发表的基于转录组数据分析可变剪接的软件。下面是官方网扎你上的介绍。
MATS is a computational tool to detect differential alternative splicing events from RNA-Seq data. The statistical model of MATS calculates the P-value and false discovery rate that the difference in the isoform ratio of a gene between two conditions exceeds a given user-defined threshold. From the RNA-Seq data, MATS can automatically detect and analyze alternative splicing events corresponding to all major types of alternative splicing patterns.
相关文献:
Shen S., Park JW., Huang J., Dittmar KA., Lu ZX., Zhou Q., Carstens RP., Xing Y. MATS: A Bayesian Framework for Flexible Detection of Differential Alternative Splicing from RNA-Seq Data. Nucleic Acids Research, 2012;40(8):e61 doi: 10.1093/nar/gkr1291
下载链接: