Pacbio数据相信大家都不陌生了,reads很长,但是错误很多而且错误分布在整条reads上而不是局部。这里给大家推荐一个工具BLASR(Basic Local Alignment with Successive Refinement )。BLASR可以讲pacbio的reads比对到比较剪辑错误比较少的序列上,譬如组装出来的contig等。
关于BLASR的算法以及相关的信息,可以参考其原文:
We describe the method BLASR (Basic Local Alignment with Successive Refinement) for mapping Single Molecule Sequencing (SMS) reads that are thousands to tens of thousands of bases long with divergence between the read and genome dominated by insertion and deletion error. We also present a combinatorial model of sequencing error that motivates why our approach is effective. The results indicate that mapping SMS reads is both highly specific and rapid.
关于安装:
BLASR的安装很简单,但是必须先安装hdf5 libraries。
使用:
这里利用BLASR把pacbio reads 比对到组装好的contig(target.fasta)上去。target.fasta.sa是target.fasta通过sawriter产生的suffix array。
blasr query.fa ./target.fasta -sa ./target.fasta.sa -bestn 40 -maxScore -500 -m 4 -nproc 24 -out target.m4 -maxLCPLength 15
在24核、48G内存的服务器上,将3G的pacbio reads比对到1000,000条contig(平均长度3500bp)上,大约需要3小时。
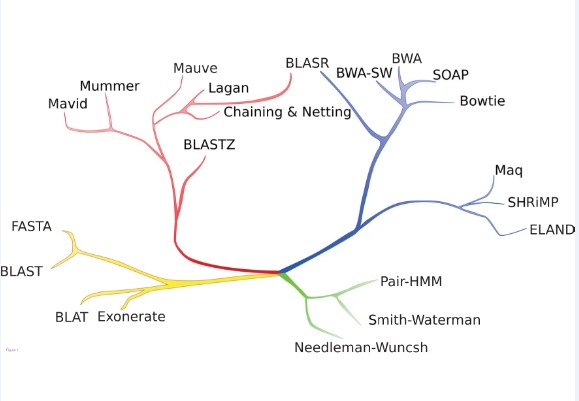
另外分享这篇论文里面比较有意思的一张图片:
图例:
Figure 1 An illustration of relationships between alignment methods.
The applications / corresponding computational restrictions shown are (green) short pairwise alignment / detailed edit model; (yellow) database search / divergent homology detection; (red) whole genome alignment / alignment of long sequences with structural rearrangements; and (blue) short read mapping / rapid alignment of massive numbers of short sequences. Although solely illustrative, methods with more similar data structures or algorithmic approaches are on closer branches. The BLASR method combines data structures from short read alignment with optimization methods from whole genome alignment