Velvet是大家所熟知的一款基于二代数据进行基因组组装的软件了,PLoB中也有多篇相关的文章来介绍velvet的安装与使用了。MetaVelvet是否听说过呢。MetaVelvet是一款基于二代数据的metagenome组装软件。下面是官方网站的相关介绍:
Introduction
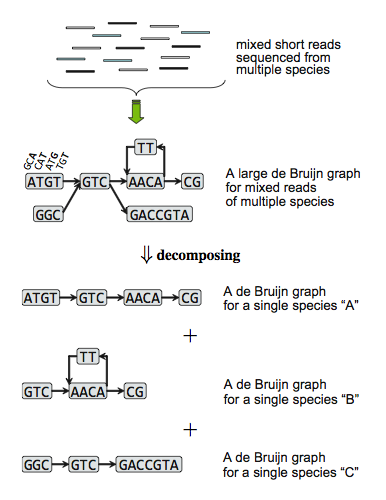
Motivation: An important step of "metagenomics" analysis is the assembly of multiple genomes from mixed sequence reads of multiple species in a microbial community. Most conventional pipelines employ a single-genome assembler with carefully optimized parameters and post-process the resulting scaffolds to correct assembly errors. Limitations of the use of a single-genome assembler for de novo metagenome assembly are that highly conserved sequences shared between different species often causes chimera contigs, and sequences of highly abundant species are likely mis-identified as repeats in a single genome.
Methods: We modified and extended a single-genome and de Bruijn-graph based assembler, Velvet, for de novo metagenome assembly. Our fundamental ideas are first decomposing de Bruijn graph constructed from mixed short reads into individual sub-graphs and second building scaffolds based on every decomposed de Bruijn sub-graph as isolate species genome.
更多关于MetaVelvet的功能、安装以及使用方法,以及下载请进官方网站:http://metavelvet.dna.bio.keio.ac.jp/