在我们分析单细胞数据的时候,需要想象力的一点就是要理解数据结构。平时我们都是如何看数据结构的呢?
library(Seurat) library(tidyverse) pbmc<-CreateSeuratObject(pbmc_small@assays$RNA@counts) pbmc%>% NormalizeData() %>% FindVariableFeatures() %>% ScaleData() %>% RunPCA() %>% FindNeighbors() %>% RunUMAP(1:10) %>% FindClusters(dims=1:0)-> pbmc pbmc An object of class Seurat 230 features across 80 samples within 1 assay Active assay: RNA (230 features) 2 dimensional reductions calculated: pca, umap
在R里面我们用的是str(...),如:
str(pbmc) Formal class 'Seurat' [package "Seurat"] with 13 slots ..@ assays :List of 1 .. ..$ RNA:Formal class 'Assay' [package "Seurat"] with 8 slots .. .. .. ..@ counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots .. .. .. .. .. ..@ i : int [1:4456] 1 5 8 11 22 30 33 34 36 38 ... .. .. .. .. .. ..@ p : int [1:81] 0 47 99 149 205 258 306 342 387 423 ... .. .. .. .. .. ..@ Dim : int [1:2] 230 80 .. .. .. .. .. ..@ Dimnames:List of 2 .. .. .. .. .. .. ..$ : chr [1:230] "MS4A1" "CD79B" "CD79A" "HLA-DRA" ... .. .. .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. .. ..@ x : num [1:4456] 1 1 3 1 1 4 1 5 1 1 ... .. .. .. .. .. ..@ factors : list() .. .. .. ..@ data :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots .. .. .. .. .. ..@ i : int [1:4456] 1 5 8 11 22 30 33 34 36 38 ... .. .. .. .. .. ..@ p : int [1:81] 0 47 99 149 205 258 306 342 387 423 ... .. .. .. .. .. ..@ Dim : int [1:2] 230 80 .. .. .. .. .. ..@ Dimnames:List of 2 .. .. .. .. .. .. ..$ : chr [1:230] "MS4A1" "CD79B" "CD79A" "HLA-DRA" ... .. .. .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. .. ..@ x : num [1:4456] 4.97 4.97 6.06 4.97 4.97 ... .. .. .. .. .. ..@ factors : list() .. .. .. ..@ scale.data : num [1:230, 1:80] -0.409 1.64 -0.428 -1.375 -0.329 ... .. .. .. .. ..- attr(*, "dimnames")=List of 2 .. .. .. .. .. ..$ : chr [1:230] "MS4A1" "CD79B" "CD79A" "HLA-DRA" ... .. .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. ..@ key : chr "rna_" .. .. .. ..@ assay.orig : NULL .. .. .. ..@ var.features : chr [1:230] "PPBP" "IGLL5" "VDAC3" "CD1C" ... .. .. .. ..@ meta.features:'data.frame': 230 obs. of 5 variables: .. .. .. .. ..$ vst.mean : num [1:230] 0.388 0.6 0.7 13.425 0.3 ... .. .. .. .. ..$ vst.variance : num [1:230] 1.025 1.281 4.365 725.463 0.871 ... .. .. .. .. ..$ vst.variance.expected : num [1:230] 1.141 2.664 4.029 745.145 0.642 ... .. .. .. .. ..$ vst.variance.standardized: num [1:230] 0.898 0.481 1.083 0.974 1.356 ... .. .. .. .. ..$ vst.variable : logi [1:230] TRUE TRUE TRUE TRUE TRUE TRUE ... .. .. .. ..@ misc : NULL ..@ meta.data :'data.frame': 80 obs. of 5 variables: .. ..$ orig.ident : Factor w/ 1 level "SeuratProject": 1 1 1 1 1 1 1 1 1 1 ... .. ..$ nCount_RNA : num [1:80] 70 85 87 127 173 70 64 72 52 100 ... .. ..$ nFeature_RNA : int [1:80] 47 52 50 56 53 48 36 45 36 41 ... .. ..$ RNA_snn_res.0.8: Factor w/ 3 levels "0","1","2": 2 2 2 2 2 2 2 2 2 2 ... .. ..$ seurat_clusters: Factor w/ 3 levels "0","1","2": 2 2 2 2 2 2 2 2 2 2 ... ..@ active.assay: chr "RNA" ..@ active.ident: Factor w/ 3 levels "0","1","2": 2 2 2 2 2 2 2 2 2 2 ... .. ..- attr(*, "names")= chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... ..@ graphs :List of 2 .. ..$ RNA_nn :Formal class 'Graph' [package "Seurat"] with 7 slots .. .. .. ..@ assay.used: chr "RNA" .. .. .. ..@ i : int [1:1600] 0 1 2 3 4 5 6 7 8 9 ... .. .. .. ..@ p : int [1:81] 0 10 17 40 57 101 124 141 153 178 ... .. .. .. ..@ Dim : int [1:2] 80 80 .. .. .. ..@ Dimnames :List of 2 .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. ..@ x : num [1:1600] 1 1 1 1 1 1 1 1 1 1 ... .. .. .. ..@ factors : list() .. ..$ RNA_snn:Formal class 'Graph' [package "Seurat"] with 7 slots .. .. .. ..@ assay.used: chr "RNA" .. .. .. ..@ i : int [1:4174] 0 1 2 3 4 5 6 7 8 9 ... .. .. .. ..@ p : int [1:81] 0 68 132 181 230 277 326 375 424 487 ... .. .. .. ..@ Dim : int [1:2] 80 80 .. .. .. ..@ Dimnames :List of 2 .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. ..@ x : num [1:4174] 1 0.6 0.6 0.6 0.538 ... .. .. .. ..@ factors : list() ..@ neighbors : list() ..@ reductions :List of 2 .. ..$ pca :Formal class 'DimReduc' [package "Seurat"] with 9 slots .. .. .. ..@ cell.embeddings : num [1:80, 1:50] 3.12 3.56 2.4 3.43 2.78 ... .. .. .. .. ..- attr(*, "dimnames")=List of 2 .. .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. .. ..$ : chr [1:50] "PC_1" "PC_2" "PC_3" "PC_4" ... .. .. .. ..@ feature.loadings : num [1:230, 1:50] 0.05711 0.00738 0.03005 -0.04766 0.05598 ... .. .. .. .. ..- attr(*, "dimnames")=List of 2 .. .. .. .. .. ..$ : chr [1:230] "PPBP" "IGLL5" "VDAC3" "CD1C" ... .. .. .. .. .. ..$ : chr [1:50] "PC_1" "PC_2" "PC_3" "PC_4" ... .. .. .. ..@ feature.loadings.projected: num[0 , 0 ] .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ global : logi FALSE .. .. .. ..@ stdev : num [1:50] 5.75 5.21 4.32 3.62 2.77 ... .. .. .. ..@ key : chr "PC_" .. .. .. ..@ jackstraw :Formal class 'JackStrawData' [package "Seurat"] with 4 slots .. .. .. .. .. ..@ empirical.p.values : num[0 , 0 ] .. .. .. .. .. ..@ fake.reduction.scores : num[0 , 0 ] .. .. .. .. .. ..@ empirical.p.values.full: num[0 , 0 ] .. .. .. .. .. ..@ overall.p.values : num[0 , 0 ] .. .. .. ..@ misc :List of 1 .. .. .. .. ..$ total.variance: num 230 .. ..$ umap:Formal class 'DimReduc' [package "Seurat"] with 9 slots .. .. .. ..@ cell.embeddings : num [1:80, 1:2] 5.07 5.31 4.72 5.06 5.45 ... .. .. .. .. ..- attr(*, "scaled:center")= num [1:2] 1.78 -8.75 .. .. .. .. ..- attr(*, "dimnames")=List of 2 .. .. .. .. .. ..$ : chr [1:80] "ATGCCAGAACGACT" "CATGGCCTGTGCAT" "GAACCTGATGAACC" "TGACTGGATTCTCA" ... .. .. .. .. .. ..$ : chr [1:2] "UMAP_1" "UMAP_2" .. .. .. ..@ feature.loadings : num[0 , 0 ] .. .. .. ..@ feature.loadings.projected: num[0 , 0 ] .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ global : logi TRUE .. .. .. ..@ stdev : num(0) .. .. .. ..@ key : chr "UMAP_" .. .. .. ..@ jackstraw :Formal class 'JackStrawData' [package "Seurat"] with 4 slots .. .. .. .. .. ..@ empirical.p.values : num[0 , 0 ] .. .. .. .. .. ..@ fake.reduction.scores : num[0 , 0 ] .. .. .. .. .. ..@ empirical.p.values.full: num[0 , 0 ] .. .. .. .. .. ..@ overall.p.values : num[0 , 0 ] .. .. .. ..@ misc : list() ..@ images : list() ..@ project.name: chr "SeuratProject" ..@ misc : list() ..@ version :Classes 'package_version', 'numeric_version' hidden list of 1 .. ..$ : int [1:3] 3 1 2 ..@ commands :List of 7 .. ..$ NormalizeData.RNA :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "NormalizeData.RNA" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:27" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "NormalizeData(.)" .. .. .. ..@ params :List of 5 .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ normalization.method: chr "LogNormalize" .. .. .. .. ..$ scale.factor : num 10000 .. .. .. .. ..$ margin : num 1 .. .. .. .. ..$ verbose : logi TRUE .. ..$ FindVariableFeatures.RNA:Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "FindVariableFeatures.RNA" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:28" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "FindVariableFeatures(.)" .. .. .. ..@ params :List of 12 .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ selection.method : chr "vst" .. .. .. .. ..$ loess.span : num 0.3 .. .. .. .. ..$ clip.max : chr "auto" .. .. .. .. ..$ mean.function :function (mat, display_progress) .. .. .. .. ..$ dispersion.function:function (mat, display_progress) .. .. .. .. ..$ num.bin : num 20 .. .. .. .. ..$ binning.method : chr "equal_width" .. .. .. .. ..$ nfeatures : num 2000 .. .. .. .. ..$ mean.cutoff : num [1:2] 0.1 8 .. .. .. .. ..$ dispersion.cutoff : num [1:2] 1 Inf .. .. .. .. ..$ verbose : logi TRUE .. ..$ ScaleData.RNA :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "ScaleData.RNA" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:28" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "ScaleData(.)" .. .. .. ..@ params :List of 10 .. .. .. .. ..$ features : chr [1:230] "PPBP" "IGLL5" "VDAC3" "CD1C" ... .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ model.use : chr "linear" .. .. .. .. ..$ use.umi : logi FALSE .. .. .. .. ..$ do.scale : logi TRUE .. .. .. .. ..$ do.center : logi TRUE .. .. .. .. ..$ scale.max : num 10 .. .. .. .. ..$ block.size : num 1000 .. .. .. .. ..$ min.cells.to.block: num 80 .. .. .. .. ..$ verbose : logi TRUE .. ..$ RunPCA.RNA :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "RunPCA.RNA" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:29" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "RunPCA(.)" .. .. .. ..@ params :List of 10 .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ npcs : num 50 .. .. .. .. ..$ rev.pca : logi FALSE .. .. .. .. ..$ weight.by.var : logi TRUE .. .. .. .. ..$ verbose : logi TRUE .. .. .. .. ..$ ndims.print : int [1:5] 1 2 3 4 5 .. .. .. .. ..$ nfeatures.print: num 30 .. .. .. .. ..$ reduction.name : chr "pca" .. .. .. .. ..$ reduction.key : chr "PC_" .. .. .. .. ..$ seed.use : num 42 .. ..$ FindNeighbors.RNA.pca :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "FindNeighbors.RNA.pca" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:29" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "FindNeighbors(.)" .. .. .. ..@ params :List of 13 .. .. .. .. ..$ reduction : chr "pca" .. .. .. .. ..$ dims : int [1:10] 1 2 3 4 5 6 7 8 9 10 .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ k.param : num 20 .. .. .. .. ..$ compute.SNN : logi TRUE .. .. .. .. ..$ prune.SNN : num 0.0667 .. .. .. .. ..$ nn.method : chr "rann" .. .. .. .. ..$ annoy.metric: chr "euclidean" .. .. .. .. ..$ nn.eps : num 0 .. .. .. .. ..$ verbose : logi TRUE .. .. .. .. ..$ force.recalc: logi FALSE .. .. .. .. ..$ do.plot : logi FALSE .. .. .. .. ..$ graph.name : chr [1:2] "RNA_nn" "RNA_snn" .. ..$ RunUMAP.RNA.pca :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "RunUMAP.RNA.pca" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:33" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "RunUMAP(., 1:10)" .. .. .. ..@ params :List of 20 .. .. .. .. ..$ dims : int [1:10] 1 2 3 4 5 6 7 8 9 10 .. .. .. .. ..$ reduction : chr "pca" .. .. .. .. ..$ assay : chr "RNA" .. .. .. .. ..$ umap.method : chr "uwot" .. .. .. .. ..$ n.neighbors : int 30 .. .. .. .. ..$ n.components : int 2 .. .. .. .. ..$ metric : chr "cosine" .. .. .. .. ..$ learning.rate : num 1 .. .. .. .. ..$ min.dist : num 0.3 .. .. .. .. ..$ spread : num 1 .. .. .. .. ..$ set.op.mix.ratio : num 1 .. .. .. .. ..$ local.connectivity : int 1 .. .. .. .. ..$ repulsion.strength : num 1 .. .. .. .. ..$ negative.sample.rate: int 5 .. .. .. .. ..$ uwot.sgd : logi FALSE .. .. .. .. ..$ seed.use : int 42 .. .. .. .. ..$ angular.rp.forest : logi FALSE .. .. .. .. ..$ verbose : logi TRUE .. .. .. .. ..$ reduction.name : chr "umap" .. .. .. .. ..$ reduction.key : chr "UMAP_" .. ..$ FindClusters :Formal class 'SeuratCommand' [package "Seurat"] with 5 slots .. .. .. ..@ name : chr "FindClusters" .. .. .. ..@ time.stamp : POSIXct[1:1], format: "2020-06-01 22:43:33" .. .. .. ..@ assay.used : chr "RNA" .. .. .. ..@ call.string: chr "FindClusters(., dims = 1:0)" .. .. .. ..@ params :List of 10 .. .. .. .. ..$ graph.name : chr "RNA_snn" .. .. .. .. ..$ modularity.fxn : num 1 .. .. .. .. ..$ resolution : num 0.8 .. .. .. .. ..$ method : chr "matrix" .. .. .. .. ..$ algorithm : num 1 .. .. .. .. ..$ n.start : num 10 .. .. .. .. ..$ n.iter : num 10 .. .. .. .. ..$ random.seed : num 0 .. .. .. .. ..$ group.singletons: logi TRUE .. .. .. .. ..$ verbose : logi TRUE ..@ tools : list()
别说看了,拉鼠标手都能拉疼。那么我们能不能基于str(pbmc)的结果做一个思维导图呢?就像这样:
如果能够这样查看,那不是美滋滋的吗?
需求有了,就差行动了,我们来找代码:
library(mindr)
(out <- capture.output(str(pbmc)))
out2 <- paste(out, collapse="n")
mm(gsub("\.\.@","# ",gsub("\.\. ","#",out2)),type ="text",root= "Seurat")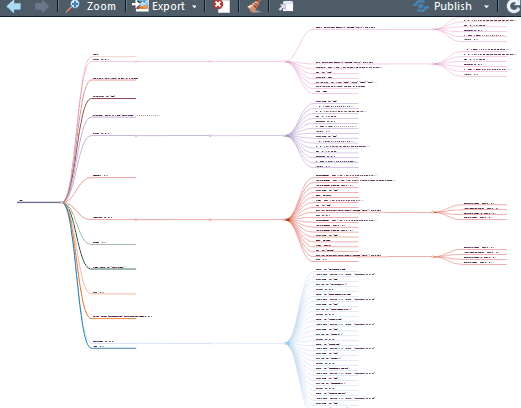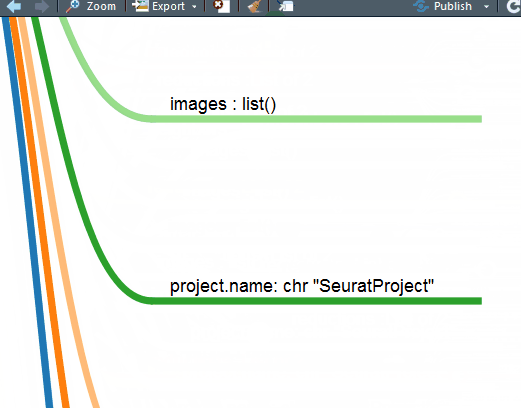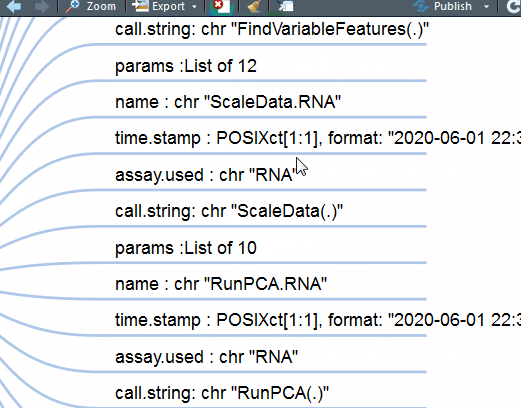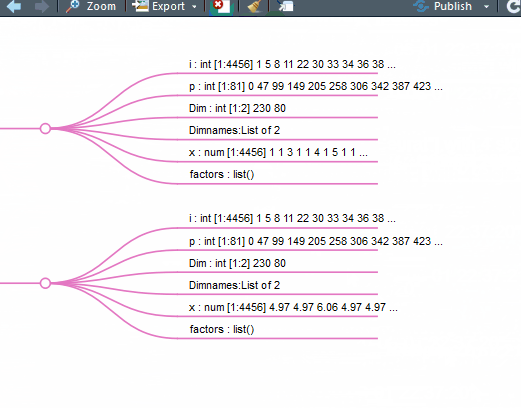
这下好了,你对单细胞Seurat数据对象做了什么一目了然。


